An S4 Class implementing Diffusion Maps
Slots
funA function that does the embedding and returns a dimRedResult object.
stdparsThe standard parameters for the function.
General usage
Dimensionality reduction methods are S4 Classes that either be used
directly, in which case they have to be initialized and a full
list with parameters has to be handed to the @fun()
slot, or the method name be passed to the embed function and
parameters can be given to the ..., in which case
missing parameters will be replaced by the ones in the
@stdpars.
Parameters
Diffusion Maps can take the following parameters:
- d
a function transforming a matrix row wise into a distance matrix or
distobject, e.g.dist.- ndim
The number of dimensions
- eps
The epsilon parameter that determines the diffusion weight matrix from a distance matrix
d, \(exp(-d^2/eps)\), if set to"auto"it will be set to the median distance to the 0.01*n nearest neighbor.- t
Time-scale parameter. The recommended value, 0, uses multiscale geometry.
- delta
Sparsity cut-off for the symmetric graph Laplacian, a higher value results in more sparsity and faster calculation. The predefined value is 10^-5.
Implementation
Wraps around diffuse, see there for
details. It uses the notation of Richards et al. (2009) which is
slightly different from the one in the original paper (Coifman and
Lafon, 2006) and there is no \(\alpha\) parameter.
There is also an out-of-sample extension, see examples.
References
Richards, J.W., Freeman, P.E., Lee, A.B., Schafer, C.M., 2009. Exploiting Low-Dimensional Structure in Astronomical Spectra. ApJ 691, 32. doi:10.1088/0004-637X/691/1/32
Coifman, R.R., Lafon, S., 2006. Diffusion maps. Applied and Computational Harmonic Analysis 21, 5-30. doi:10.1016/j.acha.2006.04.006
See also
Other dimensionality reduction methods:
DRR-class,
DrL-class,
FastICA-class,
FruchtermanReingold-class,
HLLE-class,
Isomap-class,
KamadaKawai-class,
MDS-class,
NNMF-class,
PCA-class,
PCA_L1-class,
UMAP-class,
dimRedMethod-class,
dimRedMethodList(),
kPCA-class,
nMDS-class,
tSNE-class
Examples
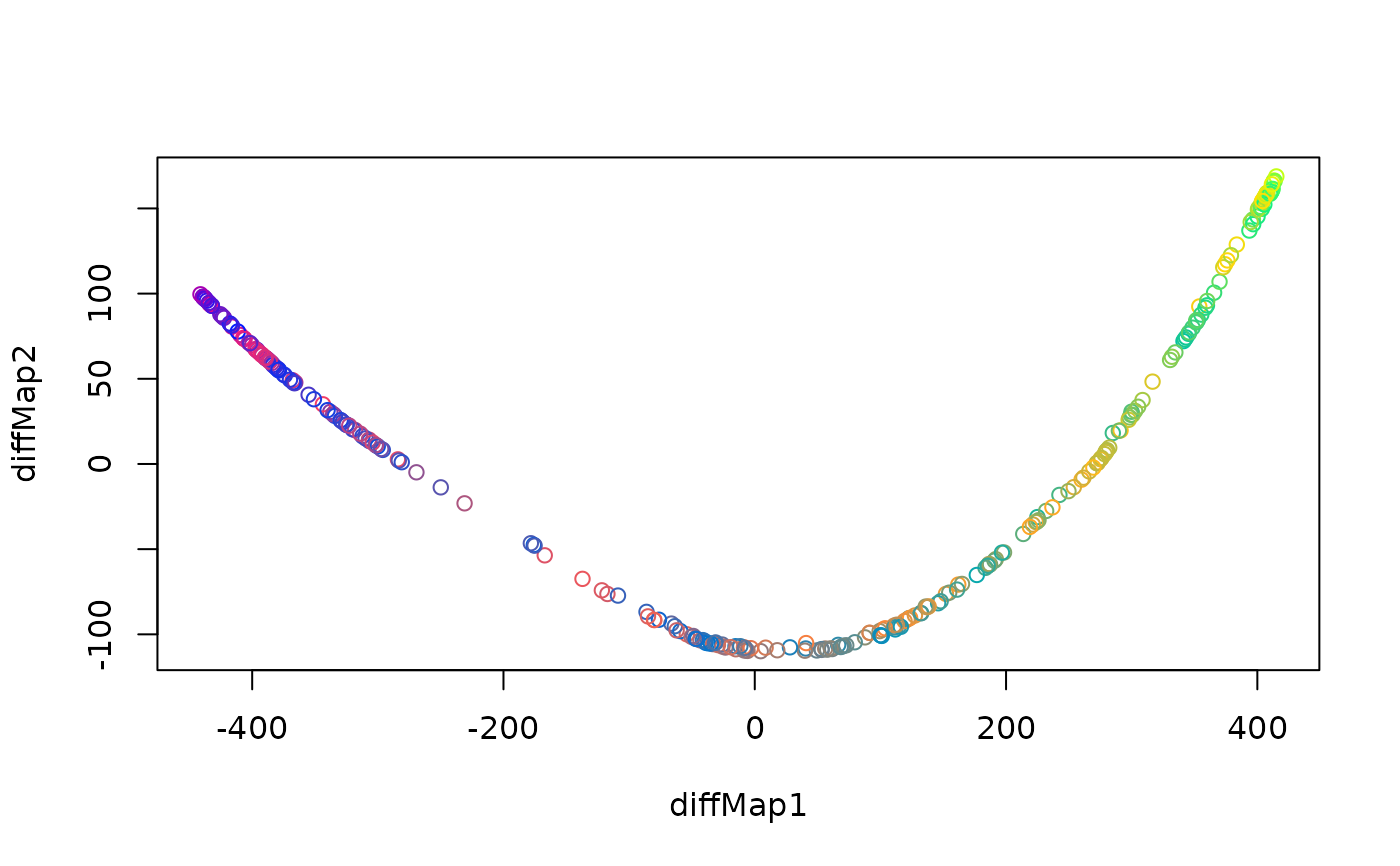
if(requireNamespace("diffusionMap", quietly = TRUE)) {
dat <- loadDataSet("3D S Curve", n = 300)
emb <- embed(dat, "DiffusionMaps")
plot(emb, type = "2vars")
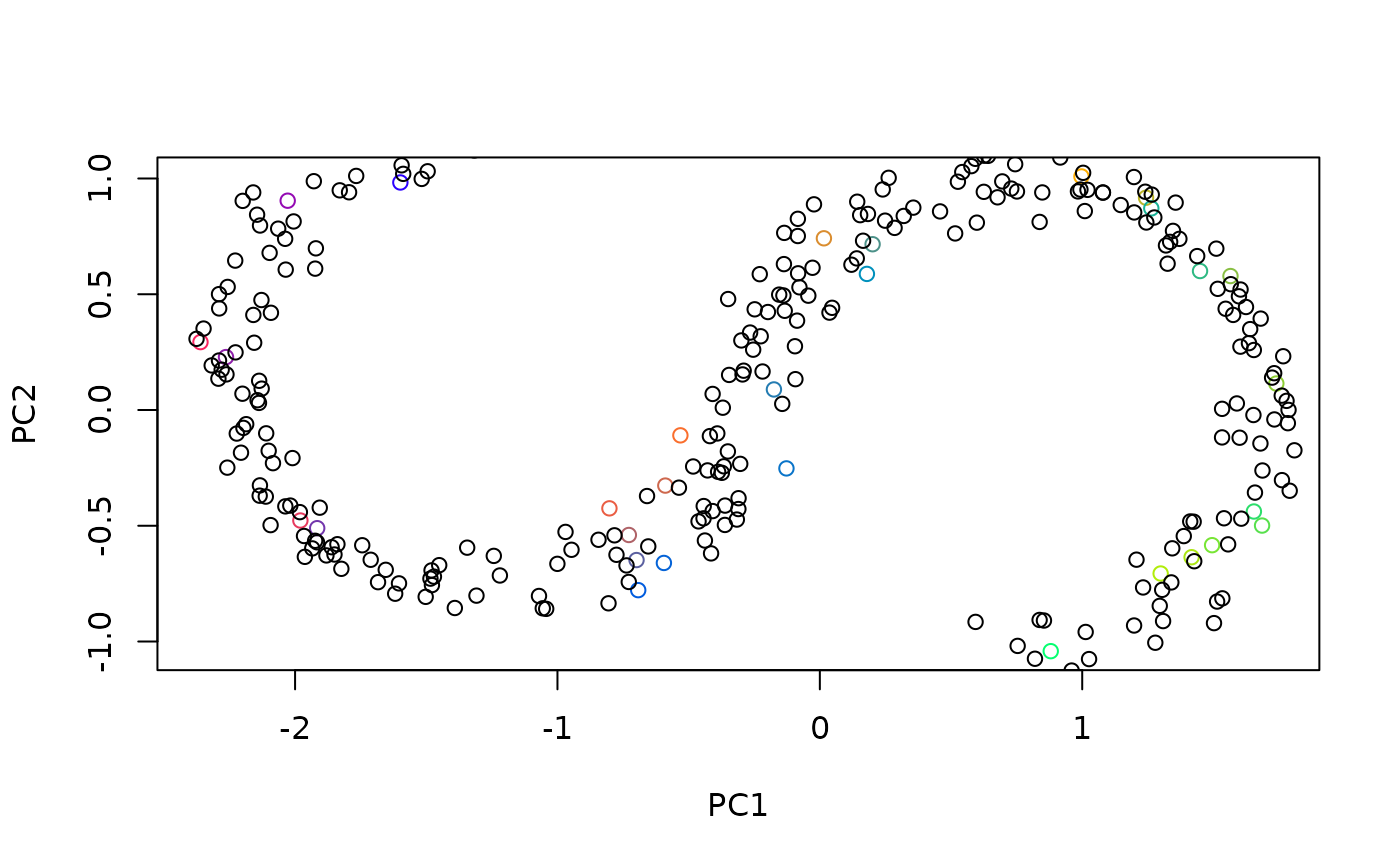
# predicting is possible:
samp <- sample(floor(nrow(dat) / 10))
emb2 <- embed(dat[samp])
emb3 <- predict(emb2, dat[-samp])
plot(emb2, type = "2vars")
points(getData(emb3))
}
#> Performing eigendecomposition
#> Computing Diffusion Coordinates
#> Elapsed time: 0.031 seconds